Scripts for Annotation
by Umer Zeeshan Ijaz and Chris Quince
Bash one-liners for extracting enzyme information from annotated GBK files
Script for extracting/drawing contigs from the GBK file. These contigs contain the enzymes we are interested in
Script for extracting all known enzyme sequences from Uniprot databases
Script for finding motifs within enzyme sequences
Scripts for searching CDS regions extracted from PROKKA against NCBI's CDD databases
We will use rest-style KEGG API to resolve enzyme details in genbank files. The API uses KEGG ENZYME database, which is an implementation of the Enzyme Nomenclature (EC Numbers) on the ExplorEnz database, and is maintained in the KEGG LIGAND relational database with additional annotation of reaction hierarchy, organism information, and sequence data links.
To use these one-liners on your genbank files, replace test.gbk with the name of the file you are using. First step is to extract a tab-separated list of only those contigs which have enzymes in them (all other contigs are ignored):
[uzi@quince-srv2 ~/annotation]$ awk '/EC_number/{print gensub(" +/EC_number=\"(.*)\"","ec:\\1","g")}/^LOCUS/{print ";"$2}' test.gbk | tr '\n' '\t' | tr ';' '\n' | awk 'NF>1{print $0}'
NODE_2925_length_923_cov_1.698808 ec:4.2.1.17
NODE_4520_length_55408_cov_7.328635 ec:1.11.1.6 ec:1.2.1.22 ec:1.1.99.1 ec:2.5.1.18
NODE_6043_length_823_cov_1.500607 ec:6.2.1.25
NODE_918_length_270_cov_10.744445 ec:1.5.3.1
NODE_312_length_2459_cov_13.763318 ec:4.3.1.17
NODE_282_length_4935_cov_13.040932 ec:2.7.6.5 ec:3.1.7.2 ec:2.7.7.6 ec:2.7.4.8
NODE_3153_length_49002_cov_8.110995 ec:1.4.3.19 ec:4.1.3.39 ec:5.1.1.8 ec:3.5.4.22 ec:1.2.1.26 ec:1.1.1.46 ec:1.1.1.1 ec:4.1.1.28
NODE_241_length_4648_cov_13.109509 ec:4.2.1.20
NODE_533_length_2973_cov_13.546249 ec:4.2.1.20
NODE_2986_length_180_cov_3.444444 ec:1.2.1.38
NODE_2957_length_228_cov_5.881579 ec:4.2.1.52
NODE_2313_length_214_cov_3.420561 ec:1.14.11.17
NODE_2177_length_8710_cov_10.954764 ec:4.1.1.36 ec:6.3.2.5 ec:3.6.1.23 ec:5.4.2.8 ec:5.4.2.2 ec:2.7.2.8 ec:2.4.2.10
NODE_2125_length_194_cov_3.505155 ec:1.2.1.27
NODE_195_length_3240_cov_14.628086 ec:4.2.1.75 ec:4.2.1.75
NODE_395_length_847_cov_14.246754 ec:2.1.1.45
NODE_263_length_1894_cov_12.784055 ec:1.8.4.11
NODE_2068_length_256_cov_5.703125 ec:6.3.2.10
NODE_5094_length_82060_cov_7.647209 ec:5.2.1.8 ec:3.4.21.92 ec:2.5.1.54 ec:3.1.1.3 ec:5.2.1.8 ec:1.1.1.30
NODE_1254_length_19746_cov_10.331156 ec:1.6.5.5 ec:1.5.1.29
NODE_4044_length_223_cov_4.789237 ec:1.1.99.1
NODE_2933_length_5310_cov_10.508098 ec:3.4.13.19
NODE_2296_length_260_cov_4.046154 ec:2.7.1.24
NODE_618_length_171_cov_3.111111 ec:2.3.1.31
NODE_4435_length_1843_cov_2.593597 ec:6.1.1.9
NODE_1478_length_868_cov_12.869816 ec:2.4.2.17
NODE_518_length_1838_cov_14.368879 ec:2.1.1.176 ec:2.1.2.9
NODE_4252_length_45804_cov_8.195944 ec:1.11.1.6 ec:1.11.1.7 ec:2.7.1.23 ec:6.2.1.3 ec:4.1.1.18 ec:2.7.7.7 ec:3.1.26.4
NODE_1912_length_207_cov_4.357488 ec:2.1.1.13
NODE_711_length_662_cov_13.104230 ec:2.7.4.7
NODE_2276_length_2332_cov_2.129074 ec:1.1.1.25
NODE_4612_length_2770_cov_2.385560 ec:4.6.1.1
NODE_4102_length_25349_cov_10.313819 ec:2.7.7.6 ec:6.3.1.8 ec:1.11.1.6
NODE_2532_length_27470_cov_9.829596 ec:4.3.1.12 ec:4.3.1.12
NODE_2478_length_31408_cov_9.062372 ec:4.2.1.70
NODE_1699_length_5660_cov_10.990990 ec:1.5.1.29
NODE_153_length_1936_cov_12.737087 ec:1.5.1.3
NODE_3727_length_13726_cov_9.037520 ec:1.1.5.2
NODE_1207_length_2434_cov_10.899754 ec:3.1.26.5
NODE_1695_length_238_cov_2.945378 ec:5.1.3.2
NODE_4703_length_3098_cov_2.432537 ec:3.2.2.20 ec:6.1.1.14
NODE_3161_length_31677_cov_10.626669 ec:1.2.1.3 ec:4.3.1.7 ec:4.3.1.7
NODE_87_length_2776_cov_13.340418 ec:2.7.7.49
NODE_495_length_2327_cov_12.959604 ec:2.5.1.18
NODE_1135_length_515_cov_12.755340 ec:6.3.2.1
NODE_2947_length_376_cov_14.313829 ec:1.4.1.13
NODE_899_length_1304_cov_13.075920 ec:3.5.1.10
NODE_4432_length_21460_cov_9.932059 ec:3.4.16.4 ec:5.4.3.8 ec:3.4.17.13
NODE_5187_length_772_cov_2.908031 ec:2.7.6.5 ec:3.1.7.2
NODE_5103_length_4544_cov_1.866417 ec:1.6.1.2 ec:1.6.1.2
NODE_4340_length_1843_cov_2.208356 ec:2.1.2.11
NODE_1053_length_980_cov_13.608163 ec:1.2.1.2
NODE_3523_length_5106_cov_10.867215 ec:4.2.1.1 ec:3.5.4.16
NODE_258_length_1705_cov_13.052199 ec:1.2.1.26
...
The following one-liner extracts the list of all enzymes found in the genbank file and uses rest-style KEGG API to generate names from EC numbers:
[uzi@quince-srv2 ~/annotation]$ for i in $(awk '/EC_number/{print gensub(" +/EC_number=\"(.*)\"","\\1","g")}' test.gbk | sort | uniq); do echo $(curl -s http://rest.kegg.jp/find/enzyme/$i) | awk '{$1=$1"\t"}1'; done
ec:1.1.1.108 carnitine 3-dehydrogenase
ec:1.11.1.15 peroxiredoxin; thioredoxin peroxidase; tryparedoxin peroxidase; alkyl hydroperoxide reductase C22; AhpC; TrxPx; TXNPx; Prx; PRDX
ec:1.1.1.158 Transferred to 1.3.1.98
ec:1.11.1.6 catalase; equilase; caperase; optidase; catalase-peroxidase; CAT
ec:1.1.1.169 2-dehydropantoate 2-reductase; 2-oxopantoate reductase; 2-ketopantoate reductase; 2-ketopantoic acid reductase; ketopantoate reductase; ketopantoic acid reductase
ec:1.11.1.7 peroxidase; lactoperoxidase; guaiacol peroxidase; plant peroxidase; Japanese radish peroxidase; horseradish peroxidase (HRP); soybean peroxidase (SBP); extensin peroxidase; heme peroxidase; oxyperoxidase; protoheme peroxidase; pyrocatechol peroxidase; scopoletin peroxidase; Coprinus cinereus peroxidase; Arthromyces ramosus peroxidase
ec:1.11.1.9 glutathione peroxidase; GSH peroxidase; selenium-glutathione peroxidase; reduced glutathione peroxidase
ec:1.1.1.205 IMP dehydrogenase; inosine-5'-phosphate dehydrogenase; inosinic acid dehydrogenase; inosinate dehydrogenase; inosine 5'-monophosphate dehydrogenase; inosine monophosphate dehydrogenase; IMP oxidoreductase; inosine monophosphate oxidoreductase ec:1.2.1.14 Transferred to 1.1.1.205
ec:1.1.1.215 gluconate 2-dehydrogenase; 2-keto-D-gluconate reductase; 2-ketogluconate reductase
ec:1.1.1.262 4-hydroxythreonine-4-phosphate dehydrogenase; NAD+-dependent threonine 4-phosphate dehydrogenase; L-threonine 4-phosphate dehydrogenase; 4-(phosphohydroxy)-L-threonine dehydrogenase; PdxA; 4-(phosphonooxy)-L-threonine:NAD+ oxidoreductase
ec:1.1.1.274 2,5-didehydrogluconate reductase (2-dehydro-D-gluconate-forming); 2,5-diketo-D-gluconate reductase (ambiguous)
ec:1.1.1.284 S-(hydroxymethyl)glutathione dehydrogenase; NAD-linked formaldehyde dehydrogenase (incorrect); formaldehyde dehydrogenase (incorrect); formic dehydrogenase (incorrect); class III alcohol dehydrogenase; ADH3; chi-ADH; FDH (incorrect); formaldehyde dehydrogenase (glutathione) (incorrect); GS-FDH (incorrect); glutathione-dependent formaldehyde dehydrogenase (incorrect); NAD-dependent formaldehyde dehydrogenase; GD-FALDH; NAD- and glutathione-dependent formaldehyde dehydrogenase
ec:1.1.1.290 4-phosphoerythronate dehydrogenase; PdxB; PdxB 4PE dehydrogenase; 4-O-phosphoerythronate dehydrogenase
ec:1.1.1.46 L-arabinose 1-dehydrogenase
ec:1.1.1.47 glucose 1-dehydrogenase [NAD(P)+]; D-glucose dehydrogenase (NAD(P)+); hexose phosphate dehydrogenase; beta-D-glucose:NAD(P)+ 1-oxidoreductase; glucose 1-dehydrogenase
...
For the extracted enzymes, we can list all the KEGG Ortholog (KO) groups each enzyme is part of, along with their detailed description. The KO system is the basis for representation for all proteins and functional RNAs that correspond to KEGG pathway nodes, BRITE hierarchy nodes, and KEGG module nodes:
[uzi@quince-srv2 ~/annotation]$ for i in $(awk '/EC_number/{print gensub(" +/EC_number=\"(.*)\"","\\1","g")}' test.gbk | sort | uniq); do echo $(curl -s http://rest.kegg.jp/link/ko/ec:$i | grep -Po '(?<=ko:).*' | sed -e "s/K/ko:K/g") | xargs -n 1 | xargs -I {} curl -s http://rest.kegg.jp/find/ko/{} | awk -v k=$i '{print "ec:"k"\t"$0}' ; done
ec:1.10.2.2 ko:K00411 UQCRFS1, RIP1, petA; ubiquinol-cytochrome c reductase iron-sulfur subunit [EC:1.10.2.2]
ec:1.1.1.1 ko:K00001 E1.1.1.1, adh; alcohol dehydrogenase [EC:1.1.1.1]
ec:1.1.1.1 ko:K00121 frmA, ADH5, adhC; S-(hydroxymethyl)glutathione dehydrogenase / alcohol dehydrogenase [EC:1.1.1.284 1.1.1.1]
ec:1.1.1.1 ko:K04072 adhE; acetaldehyde dehydrogenase / alcohol dehydrogenase [EC:1.2.1.10 1.1.1.1]
ec:1.1.1.1 ko:K11440 gbsB; choline dehydrogenase [EC:1.1.1.1]
ec:1.1.1.1 ko:K13951 ADH1_7; alcohol dehydrogenase 1/7 [EC:1.1.1.1]
ec:1.1.1.1 ko:K13952 ADH6; alcohol dehydrogenase 6 [EC:1.1.1.1]
ec:1.1.1.1 ko:K13953 adhP; alcohol dehydrogenase, propanol-preferring [EC:1.1.1.1]
ec:1.1.1.1 ko:K13954 yiaY; alcohol dehydrogenase [EC:1.1.1.1]
ec:1.1.1.1 ko:K13980 ADH4; alcohol dehydrogenase 4 [EC:1.1.1.1]
ec:1.1.1.100 ko:K00059 fabG; 3-oxoacyl-[acyl-carrier protein] reductase [EC:1.1.1.100]
ec:1.1.1.100 ko:K11610 mabA; beta-ketoacyl ACP reductase [EC:1.1.1.100]
ec:1.1.1.108 ko:K17735 lcdH, cdhA; carnitine 3-dehydrogenase [EC:1.1.1.108]
ec:1.11.1.15 ko:K03386 E1.11.1.15, PRDX, ahpC; peroxiredoxin (alkyl hydroperoxide reductase subunit C) [EC:1.11.1.15]
ec:1.11.1.15 ko:K03564 BCP, PRXQ, DOT5; peroxiredoxin Q/BCP [EC:1.11.1.15]
ec:1.11.1.15 ko:K11065 tpx; thiol peroxidase, atypical 2-Cys peroxiredoxin [EC:1.11.1.15]
ec:1.11.1.15 ko:K11185 TRYP; cytosolic tryparedoxin peroxidase, trypanosomatid typical 2-Cys peroxiredoxin [EC:1.11.1.15]
ec:1.11.1.15 ko:K11186 MTRYP; mitochondrial tryparedoxin peroxidase, trypanosomatid typical 2-Cys peroxiredoxin [EC:1.11.1.15]
ec:1.11.1.15 ko:K11187 PRDX5; peroxiredoxin 5, atypical 2-Cys peroxiredoxin [EC:1.11.1.15]
ec:1.11.1.15 ko:K11188 PRDX6; peroxiredoxin 6, 1-Cys peroxiredoxin [EC:1.11.1.7 1.11.1.15 3.1.1.-]
ec:1.11.1.15 ko:K13279 PRDX1; peroxiredoxin 1 [EC:1.11.1.15]
ec:1.11.1.15 ko:K14171 AHP1; alkyl hydroperoxide reductase 1 [EC:1.11.1.15]
ec:1.1.1.132 ko:K00066 algD; GDP-mannose 6-dehydrogenase [EC:1.1.1.132]
ec:1.1.1.133 ko:K00067 rfbD, rmlD; dTDP-4-dehydrorhamnose reductase [EC:1.1.1.133]
ec:1.11.1.5 ko:K00428 E1.11.1.5; cytochrome c peroxidase [EC:1.11.1.5]
ec:1.1.1.157 ko:K00074 paaH, hbd, fadB, mmgB; 3-hydroxybutyryl-CoA dehydrogenase [EC:1.1.1.157]
ec:1.11.1.6 ko:K03781 katE, CAT, catB, srpA; catalase [EC:1.11.1.6]
ec:1.1.1.169 ko:K00077 panE, apbA; 2-dehydropantoate 2-reductase [EC:1.1.1.169]
ec:1.11.1.7 ko:K00430 E1.11.1.7; peroxidase [EC:1.11.1.7]
ec:1.11.1.7 ko:K10788 EPX, EPO; eosinophil peroxidase [EC:1.11.1.7]
ec:1.11.1.7 ko:K11188 PRDX6; peroxiredoxin 6, 1-Cys peroxiredoxin [EC:1.11.1.7 1.11.1.15 3.1.1.-]
ec:1.11.1.7 ko:K12550 LPO; lactoperoxidase [EC:1.11.1.7]
ec:1.11.1.9 ko:K00432 E1.11.1.9; glutathione peroxidase [EC:1.11.1.9]
ec:1.1.1.205 ko:K00088 guaB; IMP dehydrogenase [EC:1.1.1.205]
ec:1.1.1.215 ko:K00090 E1.1.1.215; gluconate 2-dehydrogenase [EC:1.1.1.215]
ec:1.1.1.22 ko:K00012 UGDH, ugd; UDPglucose 6-dehydrogenase [EC:1.1.1.22]
ec:1.1.1.23 ko:K00013 hisD; histidinol dehydrogenase [EC:1.1.1.23]
ec:1.1.1.23 ko:K14152 HIS4; phosphoribosyl-ATP pyrophosphohydrolase / phosphoribosyl-AMP cyclohydrolase / histidinol dehydrogenase [EC:3.6.1.31 3.5.4.19 1.1.1.23]
ec:1.1.1.25 ko:K00014 aroE; shikimate dehydrogenase [EC:1.1.1.25]
ec:1.1.1.25 ko:K13830 ARO1; pentafunctional AROM polypeptide [EC:4.2.3.4 4.2.1.10 1.1.1.25 2.7.1.71 2.5.1.19]
ec:1.1.1.25 ko:K13832 aroDE, DHQ-SDH; 3-dehydroquinate dehydratase / shikimate dehydrogenase [EC:4.2.1.10 1.1.1.25]
ec:1.1.1.26 ko:K00015 gyaR; glyoxylate reductase [EC:1.1.1.26]
...
Similarly, for the extracted enzymes, we can also list all the known reactions these enzymes are a part of. Each reaction is identified by the R number and is linked to ortholog groups of enzymes enabling integrated analysis of genomic and chemical information.
[uzi@quince-srv2 ~/annotation]$ for i in $(awk '/EC_number/{print gensub(" +/EC_number=\"(.*)\"","\\1","g")}' test.gbk | sort | uniq); do echo $(curl -s http://rest.kegg.jp/link/reaction/ec:$i | grep -Po '(?<=rn:).*') | xargs -n 1 | xargs -I {} curl -s http://rest.kegg.jp/find/reaction/rn:{} | awk -v k=$i '{print "ec:"k"\t"$0}' ; done
ec:1.10.2.2 rn:R02161 Ubiquinol:ferricytochrome-c oxidoreductase; Ubiquinol + 2 Ferricytochrome c <=> Ubiquinone + 2 Ferrocytochrome c + 2 H+
ec:1.1.1.1 rn:R00228 acetaldehyde:NAD+ oxidoreductase (CoA-acetylating); Acetaldehyde + CoA + NAD+ <=> Acetyl-CoA + NADH + H+
ec:1.1.1.1 rn:R00623 primary_alcohol:NAD+ oxidoreductase; Primary alcohol + NAD+ <=> Aldehyde + NADH + H+
ec:1.1.1.1 rn:R00624 Secondary_alcohol:NAD+ oxidoreductase; Secondary alcohol + NAD+ <=> Ketone + NADH + H+
ec:1.1.1.1 rn:R00754 ethanol:NAD+ oxidoreductase; Ethanol + NAD+ <=> Acetaldehyde + NADH + H+
ec:1.1.1.1 rn:R01172 butanal:NAD+ oxidoreductase (CoA-acylating); Butanal + CoA + NAD+ <=> Butanoyl-CoA + NADH + H+
ec:1.1.1.1 rn:R02124 retinol:NAD+ oxidoreductase; Retinol + NAD+ <=> Retinal + NADH + H+
ec:1.1.1.1 rn:R02878 1-Octanol:NAD+ oxidoreductase; 1-Octanol + NAD+ <=> 1-Octanal + NADH + H+
ec:1.1.1.1 rn:R04805 3alpha,7alpha,26-Trihydroxy-5beta-cholestane + NAD+ <=> 3alpha,7alpha-Dihydroxy-5beta-cholestan-26-al + NADH + H+
ec:1.1.1.1 rn:R04880 3,4-dihydroxyphenylethyleneglycol:NAD+ oxidoreductase; 3,4-Dihydroxyphenylethyleneglycol + NAD+ <=> 3,4-Dihydroxymandelaldehyde + NADH + H+
ec:1.1.1.1 rn:R05233 trans-3-Chloro-2-propene-1-ol:NAD+ oxidoreductase; trans-3-Chloro-2-propene-1-ol + NAD+ <=> trans-3-Chloroallyl aldehyde + NADH + H+
ec:1.1.1.1 rn:R05234 cis-3-chloro-2-propene-1-ol:NAD+ oxidoreductase; cis-3-Chloro-2-propene-1-ol + NAD+ <=> cis-3-Chloroallyl aldehyde + NADH + H+
ec:1.1.1.1 rn:R06917 1-hydroxymethylnaphthalene:NAD+ oxidoreductase; 1-Hydroxymethylnaphthalene + NAD+ <=> 1-Naphthaldehyde + NADH + H+
ec:1.1.1.1 rn:R06927 (2-naphthyl)methanol:NAD+ oxidoreductase; (2-Naphthyl)methanol + NAD+ <=> 2-Naphthaldehyde + NADH + H+
ec:1.1.1.1 rn:R06983 S-(hydroxymethyl)glutathione dehydrogenase; S-(Hydroxymethyl)glutathione + NAD+ <=> S-Formylglutathione + NADH + H+
ec:1.1.1.1 rn:R07105 trichloroethanol:NAD+ oxidoreductase; Chloral hydrate + NADH + H+ <=> Trichloroethanol + NAD+ + H2O
ec:1.1.1.1 rn:R07326 alcohol:NAD+ oxidoreductase; Alcohol + NAD+ <=> Aldehyde + NADH + H+
ec:1.1.1.1 rn:R07327 alcohol:NAD+ oxidoreductase; Alcohol + NAD+ <=> Ketone + NADH + H+
ec:1.1.1.1 rn:R08281 alcophosphamide:NAD+ oxidoreductase; Aldophosphamide + NADH + H+ <=> Alcophosphamide + NAD+
ec:1.1.1.1 rn:R08306 2-phenyl-1,3-propanediol monocarbamate:NAD+ oxidoreductase; 2-Phenyl-1,3-propanediol monocarbamate + NAD+ <=> 3-Carbamoyl-2-phenylpropionaldehyde + NADH + H+
ec:1.1.1.1 rn:R08310 4-hydroxy-5-phenyltetrahydro-1,3-oxazin-2-one:NAD+ oxidoreductase; 4-Hydroxy-5-phenyltetrahydro-1,3-oxazin-2-one + NAD+ <=> 5-Phenyl-1,3-oxazinane-2,4-dione + NADH + H+
ec:1.1.1.1 rn:R08557 Choline + NAD+ <=> Betaine aldehyde + NADH + H+
ec:1.1.1.1 rn:R08558 Choline + NADP+ <=> Betaine aldehyde + NADPH + H+
ec:1.1.1.100 rn:R02767 (3R)-3-Hydroxyacyl-[acyl-carrier-protein]:NADP+ oxidoreductase; (3R)-3-Hydroxyacyl-[acyl-carrier protein] + NADP+ <=> 3-Oxoacyl-[acyl-carrier protein] + NADPH + H+
ec:1.1.1.100 rn:R04533 (3R)-3-Hydroxybutanoyl-[acyl-carrier protein]:NADP+ oxidoreductase; (3R)-3-Hydroxybutanoyl-[acyl-carrier protein] + NADP+ <=> Acetoacetyl-[acp] + NADPH + H+
ec:1.1.1.100 rn:R04534 (3R)-3-Hydroxydecanoyl-[acyl-carrier-protein]:NADP+ oxidoreductase; (3R)-3-Hydroxydecanoyl-[acyl-carrier protein] + NADP+ <=> 3-Oxodecanoyl-[acp] + NADPH
ec:1.1.1.100 rn:R04536 (3R)-3-Hydroxyoctanoyl-[acyl-carrier-protein]:NADP+ oxidoreductase; (3R)-3-Hydroxyoctanoyl-[acyl-carrier protein] + NADP+ <=> 3-Oxooctanoyl-[acp] + NADPH + H+
ec:1.1.1.100 rn:R04543 (3R)-3-Hydroxypalmitoyl-[acyl-carrier-protein]:NADP+ oxidoreductase; (3R)-3-Hydroxypalmitoyl-[acyl-carrier protein] + NADP+ <=> 3-Oxohexadecanoyl-[acp] + NADPH + H+
ec:1.1.1.100 rn:R04566 (3R)-3-Hydroxytetradecanoyl-[acyl-carrier-protein]:NADP+ oxidoreductase; (3R)-3-Hydroxytetradecanoyl-[acyl-carrier protein] + NADP+ <=> 3-Oxotetradecanoyl-[acp] + NADPH + H+
ec:1.1.1.100 rn:R04953 (3R)-3-Hydroxyhexanoyl-[acyl-carrier-protein]:NADP+ oxidoreductase; (R)-3-Hydroxyhexanoyl-[acp] + NADP+ <=> 3-Oxohexanoyl-[acp] + NADPH + H+
ec:1.1.1.100 rn:R04964 (3R)-3-Hydroxydodecanoyl-[acyl-carrier-protein]:NADP+ oxidoreductase; (R)-3-Hydroxydodecanoyl-[acp] + NADP+ <=> 3-Oxododecanoyl-[acp] + NADPH + H+
ec:1.1.1.100 rn:R07759 3-Oxostearoyl-CoA + NADPH + H+ <=> 3-Hydroxyoctadecanoyl-CoA + NADP+
ec:1.1.1.100 rn:R07763 3-Oxostearoyl-[acp] + NADPH + H+ <=> 3-Hydroxyoctadecanoyl-[acp] + NADP+
ec:1.1.1.100 rn:R10116 3-hydroxyglutaryl-[acp]-methyl-ester:NADP+ oxidoreductase; 3-Ketoglutaryl-[acp] methyl ester + NADPH + H+ <=> 3-Hydroxyglutaryl-[acp] methyl ester + NADP+
ec:1.1.1.100 rn:R10120 3-hydroxypimeloyl-[acp]-methyl-ester:NADP+ oxidoreductase; 3-Ketopimeloyl-[acp] methyl ester + NADPH + H+ <=> 3-Hydroxypimeloyl-[acp] methyl ester + NADP+
ec:1.1.1.108 rn:R02395 Carnitine:NAD+ 3-oxidoreductase; Carnitine + NAD+ <=> 3-Dehydrocarnitine + NADH + H+
ec:1.11.1.15 rn:R00602 methanol:hydrogen-peroxide oxidoreductase; Methanol + Hydrogen peroxide <=> Formaldehyde + 2 H2O
ec:1.11.1.15 rn:R00698 L-phenylalanine:oxygen oxidoreductase (decarboxylating); L-Phenylalanine + Oxygen <=> 2-Phenylacetamide + CO2
ec:1.11.1.15 rn:R02596 Donor:hydrogen-peroxide oxidoreductase; Coniferyl alcohol <=> Guaiacyl lignin
ec:1.11.1.15 rn:R03919 Donor:hydrogen-peroxide oxidoreductase; Sinapyl alcohol <=> Syringyl lignin
ec:1.11.1.15 rn:R04007 Donor:hydrogen-peroxide oxidoreductase; 4-Coumaryl alcohol <=> p-Hydroxyphenyl lignin
ec:1.11.1.15 rn:R07180 thiol-containing-reductant:hydroperoxide oxidoreductase; ROOH + 2 Thiol-containing reductant <=> Alcohol + H2O + Oxidized thiol-containing reductant
ec:1.11.1.15 rn:R07443 donor:hydrogen-peroxide oxidoreductase; 5-Hydroxyconiferyl alcohol <=> 5-Hydroxy-guaiacyl lignin
ec:1.11.1.15 rn:R08360 tryparedoxin:hydroperoxide oxidoreductase; Tryparedoxin + ROOH <=> Tryparedoxin disulfide + H2O + ROH
...
We may also be interested in knowing which other organisms contain the same enzymes:
[uzi@quince-srv2 ~/annotation]$ for i in $(awk '/EC_number/{print gensub(" +/EC_number=\"(.*)\"","\\1","g")}' test.gbk | sort | uniq); do echo $(curl -s http://rest.kegg.jp/link/genome/ec:$i | grep -Po '(?<=genome:).*') | xargs -n 1 | xargs -I {} curl -s http://rest.kegg.jp/find/genome/genome:{} | awk -v k=$i '{print "ec:"k"\t"$0}' ; done
ec:1.10.2.2 genome:T01003 rno, RAT, 10116; Rattus norvegicus (Norway rat)
ec:1.10.2.2 genome:T01008 bta, BOVIN, 9913; Bos taurus (cow)
ec:1.1.1.1 genome:T00030 dme, DROME, 7227; Drosophila melanogaster (fruit fly)
ec:1.1.1.1 genome:T01001 hsa, HUMAN, 9606; Homo sapiens (human)
ec:1.1.1.1 genome:T01003 rno, RAT, 10116; Rattus norvegicus (Norway rat)
ec:1.1.1.1 genome:T01058 ecb, HORSE, 9796; Equus caballus (horse)
ec:1.1.1.100 genome:T00007 eco, ECOLI, 511145; Escherichia coli K-12 MG1655
ec:1.1.1.108 genome:T00035 pae, PSEAE, 208964; Pseudomonas aeruginosa PAO1
ec:1.11.1.15 genome:T01001 hsa, HUMAN, 9606; Homo sapiens (human)
ec:1.11.1.15 genome:T01003 rno, RAT, 10116; Rattus norvegicus (Norway rat)
ec:1.11.1.5 genome:T00005 sce, YEAST, 559292; Saccharomyces cerevisiae S288c
ec:1.1.1.157 genome:T00564 ckl, CLOK5, 431943; Clostridium kluyveri DSM 555
ec:1.11.1.6 genome:T00115 lpl, LACPL, 220668; Lactobacillus plantarum WCFS1
ec:1.11.1.6 genome:T01001 hsa, HUMAN, 9606; Homo sapiens (human)
ec:1.1.1.169 genome:T00005 sce, YEAST, 559292; Saccharomyces cerevisiae S288c
ec:1.1.1.169 genome:T00007 eco, ECOLI, 511145; Escherichia coli K-12 MG1655
ec:1.11.1.7 genome:T01008 bta, BOVIN, 9913; Bos taurus (cow)
ec:1.11.1.9 genome:T01008 bta, BOVIN, 9913; Bos taurus (cow)
ec:1.1.1.205 genome:T00566 kpn, KLEP7, 272620; Klebsiella pneumoniae subsp. pneumoniae MGH 78578
ec:1.1.1.22 genome:T01008 bta, BOVIN, 9913; Bos taurus (cow)
ec:1.1.1.23 genome:T00007 eco, ECOLI, 511145; Escherichia coli K-12 MG1655
ec:1.1.1.23 genome:T00065 stm, SALTY, 99287; Salmonella enterica subsp. enterica serovar Typhimurium LT2 (Salmonella typhimurium LT2)
ec:1.1.1.25 genome:T00005 sce, YEAST, 559292; Saccharomyces cerevisiae S288c
ec:1.1.1.25 genome:T00007 eco, ECOLI, 511145; Escherichia coli K-12 MG1655
ec:1.1.1.25 genome:T00566 kpn, KLEP7, 272620; Klebsiella pneumoniae subsp. pneumoniae MGH 78578
ec:1.1.1.262 genome:T00007 eco, ECOLI, 511145; Escherichia coli K-12 MG1655
ec:1.1.1.28 genome:T00115 lpl, LACPL, 220668; Lactobacillus plantarum WCFS1
ec:1.1.1.284 genome:T00005 sce, YEAST, 559292; Saccharomyces cerevisiae S288c
ec:1.1.1.284 genome:T00440 pde, PARDP, 318586; Paracoccus denitrificans PD1222
ec:1.1.1.284 genome:T01001 hsa, HUMAN, 9606; Homo sapiens (human)
ec:1.1.1.284 genome:T01002 mmu, MOUSE, 10090; Mus musculus (house mouse)
ec:1.1.1.290 genome:T00007 eco, ECOLI, 511145; Escherichia coli K-12 MG1655
ec:1.1.1.3 genome:T00005 sce, YEAST, 559292; Saccharomyces cerevisiae S288c
ec:1.1.1.3 genome:T00007 eco, ECOLI, 511145; Escherichia coli K-12 MG1655
ec:1.1.1.30 genome:T01003 rno, RAT, 10116; Rattus norvegicus (Norway rat)
ec:1.1.1.31 genome:T01009 ssc, PIG, 9823; Sus scrofa (pig)
ec:1.1.1.35 genome:T01003 rno, RAT, 10116; Rattus norvegicus (Norway rat)
ec:1.1.1.35 genome:T01008 bta, BOVIN, 9913; Bos taurus (cow)
ec:1.1.1.35 genome:T01009 ssc, PIG, 9823; Sus scrofa (pig)
ec:1.1.1.42 genome:T00011 afu, ARCFU, 224325; Archaeoglobus fulgidus DSM 4304 (VC-16)
ec:1.1.1.42 genome:T00054 sso, SULSO, 273057; Sulfolobus solfataricus P2
ec:1.1.1.42 genome:T01003 rno, RAT, 10116; Rattus norvegicus (Norway rat)
ec:1.1.1.42 genome:T01009 ssc, PIG, 9823; Sus scrofa (pig)
ec:1.1.1.42 genome:T01012 tcr, TRYCR, 353153; Trypanosoma cruzi CL Brener
ec:1.1.1.44 genome:T00007 eco, ECOLI, 511145; Escherichia coli K-12 MG1655
ec:1.1.1.44 genome:T00010 bsu, BACSU, 224308; Bacillus subtilis subsp. subtilis 168
ec:1.1.1.44 genome:T01003 rno, RAT, 10116; Rattus norvegicus (Norway rat)
ec:1.1.1.47 genome:T01008 bta, BOVIN, 9913; Bos taurus (cow)
ec:1.1.1.47 genome:T01009 ssc, PIG, 9823; Sus scrofa (pig)
ec:1.1.1.49 genome:T00005 sce, YEAST, 559292; Saccharomyces cerevisiae S288c
ec:1.1.1.49 genome:T00007 eco, ECOLI, 511145; Escherichia coli K-12 MG1655
ec:1.1.1.49 genome:T00013 aae, AQUAE, 224324; Aquifex aeolicus VF5
ec:1.1.1.49 genome:T00022 tma, THEMA, 243274; Thermotoga maritima MSB8
ec:1.1.1.49 genome:T01001 hsa, HUMAN, 9606; Homo sapiens (human)
ec:1.1.1.49 genome:T01009 ssc, PIG, 9823; Sus scrofa (pig)
ec:1.1.1.49 genome:T01013 tbr, 999953; Trypanosoma brucei brucei 927/4 GUTat10.1
ec:1.1.1.49 genome:T01034 ncr, 367110; Neurospora crassa OR74A
...
To extract/draw contigs that match a particular enzyme you are interested in, use the genbank_enzymes.py script. The usage information is as follows:
[uzi@quince-srv2 ~/annotation]$ python genbank_enzymes.py
Usage:
python genbank_enzymes.py -g <gbk_file> -o <output_folder> -l <list_file> [OPTIONS]
Options:
-s (--font_size) Size of labels on diagrams (Default:12)
-f (--fragments) Number of fragments to split the genome diagram (Default:1)
-n (--name_length) Maximum length of names for output files (Default:10)
Say we are interested in extracting only those contigs that contain the two enzymes provided in IDs.txt:
[uzi@quince-srv2 ~/annotation]$ cat IDs.txt
catalase
1.1.1.22
We run the script as follows on our test.gbk file as follows:
[uzi@quince-srv2 ~/annotation]$ python genbank_enzymes.py -g test.gbk -o test -l IDs.txt -s 8 -n 9
Found catalase in NODE_4520_length_55408_cov_7.328635
Generated test/NODE_4520.gbk, test/NODE_4520.fasta, and test/NODE_4520.pdf
Found catalase in NODE_3283_length_187584_cov_8.268562
Generated test/NODE_3283.gbk, test/NODE_3283.fasta, and test/NODE_3283.pdf
Found catalase in NODE_4252_length_45804_cov_8.195944
Generated test/NODE_4252.gbk, test/NODE_4252.fasta, and test/NODE_4252.pdf
Found catalase in NODE_4102_length_25349_cov_10.313819
Generated test/NODE_4102.gbk, test/NODE_4102.fasta, and test/NODE_4102.pdf
Found catalase in NODE_1605_length_312_cov_4.407051
Generated test/NODE_1605.gbk, test/NODE_1605.fasta, and test/NODE_1605.pdf
Found 1.1.1.22 in NODE_5958_length_795089_cov_7.501846
Generated test/NODE_5958.gbk, test/NODE_5958.fasta, and test/NODE_5958.pdf
Found catalase in NODE_854_length_21339_cov_11.382820
Generated test/NODE_854_.gbk, test/NODE_854_.fasta, and test/NODE_854_.pdf
Found 1.1.1.22 in NODE_5900_length_61310_cov_7.799576
Generated test/NODE_5900.gbk, test/NODE_5900.fasta, and test/NODE_5900.pdf
Note that the script also generates annotated diagrams for contigs with enzymes on them. On these diagrams, hypothetical proteins are drawn as orange boxes, any other CDS with known information as light blue boxes, enzymes that match the EC numbers/names provided in the list as red arrows, and other enzymes as green arrows, respectively.
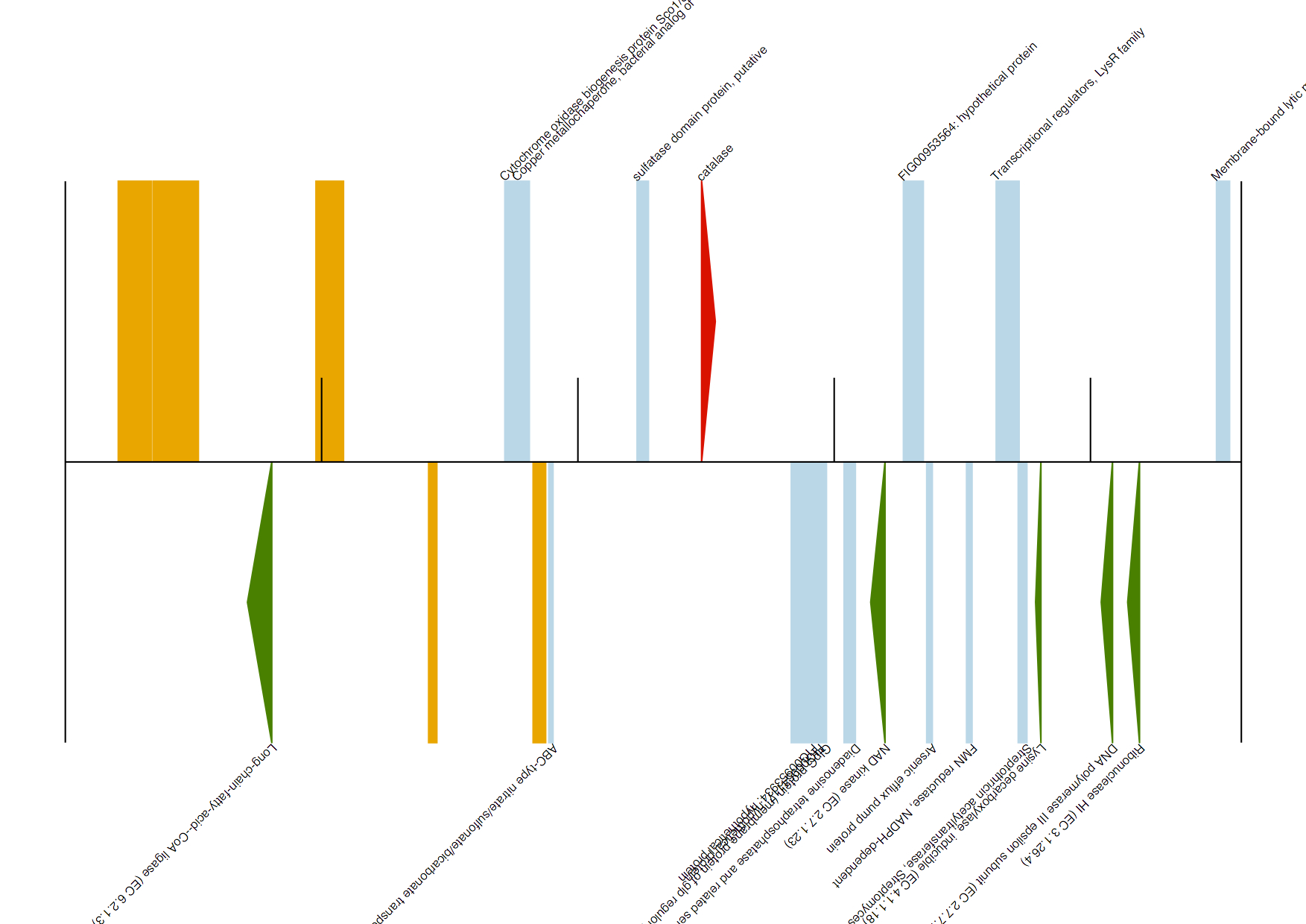
NODE_4252.pdf
|
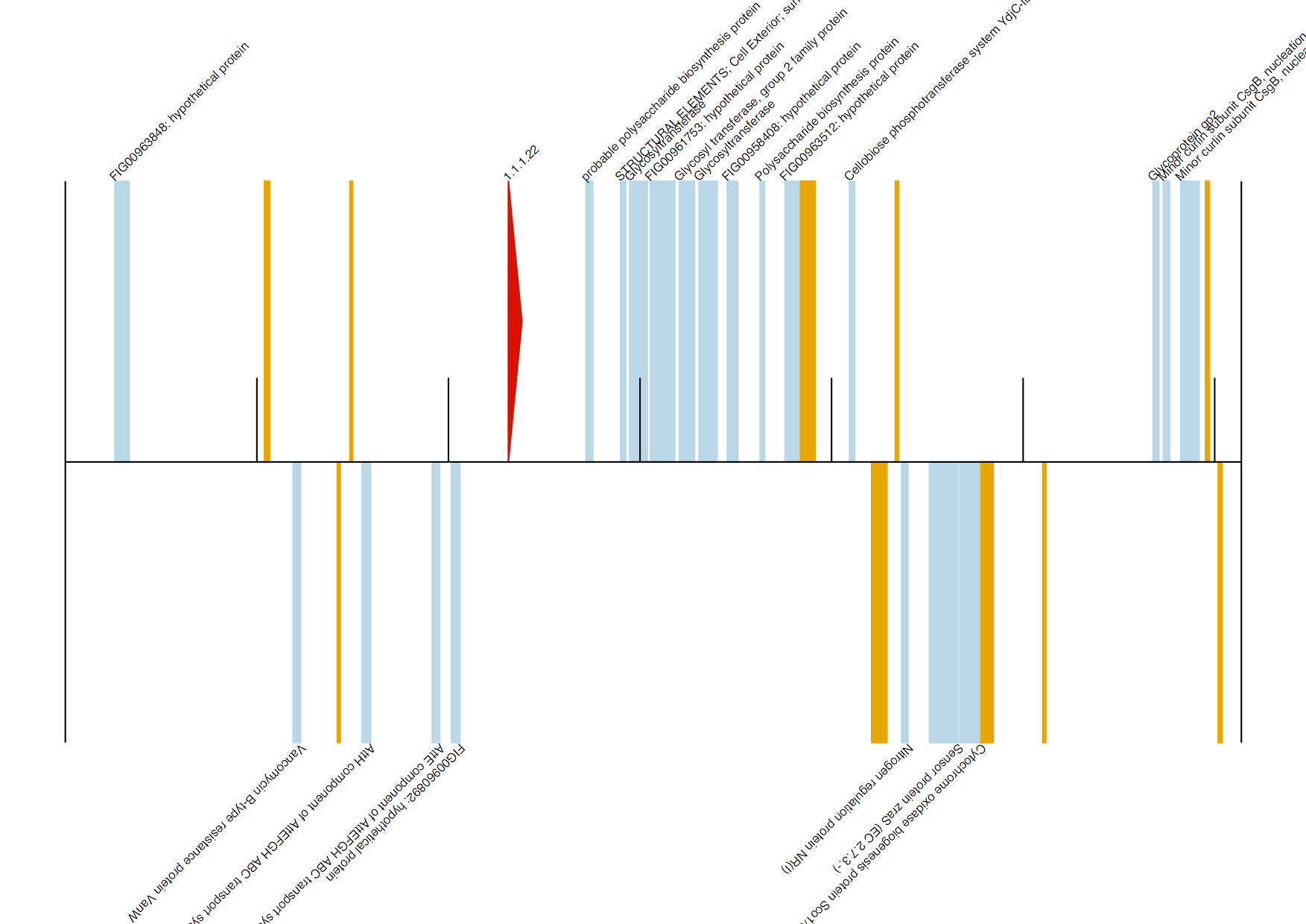
NODE_5900.pdf
|
There are two types of databases available at ftp.expasy.org: uniprot_sprot.dat.gz that is manually curated and reviewed; and uniprot_trmbl.dat.gz that is automatically curated but not reviewed. These databases are the backend for what you see on www.uniprot.org.
You can download these databases and can extract records from them using the extract_fasta_swissprot.py script. One of the output header formats can also give you taxonomy IDs to help you place the sequences on the phylogenetic tree. Please note that the search pattern is case-sensitive. The usage information is as follows:
[uzi@quince-srv2 ~/annotation]$ python extract_fasta_swissprot.py --help
Example usages:
python extract_fasta_swissprot.py -i uniprot_sprot.dat.gz -p "ATP16" -f 5 -m 4 > ATP16.fasta
python extract_fasta_swissprot.py -i uniprot_sprot.dat.gz -p "Acetyl-CoA hydrolase" > Acetyl_CoA_hydrolase.fasta
python extract_fasta_swissprot.py -i uniprot_sprot.dat.gz -p "Candida albicans" -f 2 > Candida_albicans.fasta
Parameters:
-i (--input) Compressed SwissProt file
-p (--pattern) Search pattern
-m (--format) Header format
Available search types:
-f 1 Protein Names (including Alternative Name) [DEFAULT]
-f 2 Organism Name
-f 3 Keywords
-f 4 Comments
-f 5 Gene Name
-f 6 Accession Number
-f 7 Organism Classification
Available header formats:
-m 1 >Accession IDs | Organism Name [DEFAULT]
-m 2 >Accesion IDs | Gene Name | Organism Name
-m 3 >Entry ID
-m 4 >Entry ID | Taxonomy IDs
For example, you can find all the Acetyl-CoA hydrolases (EC 3.1.2.1) using:
[uzi@quince-srv2 ~/annotation]$ python extract_fasta_swissprot.py -i uniprot_sprot.dat.gz -p "Acetyl-CoA hydrolase" > Acetyl_CoA_hydrolase.fasta
You can then align all the sequences using ClustalW. The alignment file generated for the above Acetyl_CoA_hydrolase.fasta is as follows:
CLUSTAL 2.1 multiple sequence alignment
P83773_Q59YK9|Candida_albicans --------MSAILKQRVKYAPYLKKLRT-GEQCIDLF--KHGQYLGWSGF 39
Q6BKW1|Debaryomyces_hansenii --------MSSILKQRVRYAPYLQKLRT-AEQCVELF--KHGQYLGWSGF 39
Q6C3Z9|Yarrowia_lipolytica --------MSAVLKQRIRYKPYLDKIRT-AAQCVDLFGAKPNQYIGWSGF 41
Q6FPF3|Candida_glabrata ------MTVSKLLKERVRYAPYLAKVKT-PEELIPLF--KNGQYLGWSGF 41
P32316_D6VPY5|Saccharomyces_ce ------MTISNLLKQRVRYAPYLKKVKE-AHELIPLF--KNGQYLGWSGF 41
Q6CNR2|Kluyveromyces_lactis ------MTVSRLLKDRVRYAPYLKKVKP-VEELIPLF--KDGQYIGWSGF 41
Q754Q2|Ashbya_gossypii ------MTVSQLLKQRVRYAPYLSKVRR-AEELLPLF--KHGQYIGWSGF 41
Q9UUJ9_P78872|Schizosaccharomy --------MPSLLSSRIRNAEFAKKIVTDPAKLVPYF--KNGDYVGWSGF 40
P15937_Q7RV87|Neurospora_crass ---MASPIASAALRARVKRPSMLKKLCN-PEDMLQHF--PNGAYIGWSGF 44
Q54K91|Dictyostelium_discoideu MHKIVQQGSKSLIESRIKRKSLLSKVVTDVSQLIPYF--QNGHYVSMGGF 48
: *:: *: . : * . *:. .**
P83773_Q59YK9|Candida_albicans TGVGAPKVIPTTLVDHVEKNNLQG--KLGFHLFVGASAGPE-ESRWAENN 86
Q6BKW1|Debaryomyces_hansenii TGVGAPKVVPDAIADHVEKNNLQG--KLGFNLFVGASAGPE-EGRWAEND 86
Q6C3Z9|Yarrowia_lipolytica TGVGAPKVVPDAVSKHVEENNLQGHENWRYNLFVGASAGMEIESRWANNN 91
Q6FPF3|Candida_glabrata TGVGTPKAVPDALIKHVEDNNLQG--KLRFNLFVGASAGPE-ENKWAEHD 88
P32316_D6VPY5|Saccharomyces_ce TGVGTPKAVPEALIDHVEKNNLQG--KLRFNLFVGASAGPE-ENRWAEHD 88
Q6CNR2|Kluyveromyces_lactis TGVGAPKAVPEALIKHVEENNLQG--KLRFNLFVGASAGPE-ECKWAEHD 88
Q754Q2|Ashbya_gossypii TGVGAPKVIPTALADHVEKNGLQG--QLAFNLFVGASAGPE-ENRWADLD 88
Q9UUJ9_P78872|Schizosaccharomy TGVGYPKMIPNALARHVEENNLQG--KLRFKLFVGASGGADTECKWAELN 88
P15937_Q7RV87|Neurospora_crass TGVGYPKKVPTMLADHVEKNGLQG--QLKYSLFVGASAGAETENRWAALD 92
Q54K91|Dictyostelium_discoideu AGTGYPKVVPIALADHVEKNGLQG--KLKMNLFVGASVGPETEDRWAMLD 96
As another example, you can find all the proteins for the organism Candida albicans as:
[uzi@quince-srv2 ~/annotation]$ python extract_fasta_swissprot.py -i uniprot_sprot.dat.gz -p "Candida albicans" -f 2 > Candida_albicans.fasta
The FASTA file will have the following records:
>O42766:Q5AI38|Candida_albicans
MPASREDSVYLAKLAEQAERYEEMVENMKAVASSGQELSVEERNLLSVAYKNVIGARRAS
WRIVSSIEQKEEAKGNESQVALIRDYRAKIEAELSKICEDILSVLSDHLITSAQTGESKV
FYYKMKGDYHRYLAEFAIAEKRKEAADLSLEAYKAASDVAVTELPPTHPIRLGLALNFSV
FYYEILNSPDRACHLAKQAFDDAVADLETLSEDSYKDSTLIMQLLRDNLTLWTDLSEAPA
ATEEQQQSSQAPAAQPTEGKADQE
>Q59Z17:Q59Z79|Candida_albicans
MVKKVLLINGPNLNLLGTREPEKYGTTSLSDIEQAAIEQAKLKNNDSEVLAFQSNTEGFI
IDRIHEAKRQGVGFVVINAGAYTHTSVGIRDALLGTAIPFIEVHITNVHQREPFRHQSYL
SDKAVAVICGLGVYGYTAAIEYALNY
>Q59K86|Candida_albicans
MVLGQPINIIKWIEENGDLLKPPVNNFCLHRGGFTIMIVGGPNERSDYHINQTPEYFYQF
KGTMCLKVVDDGEFKDIFINEGDSFLLPPNVPHNPCRYENTIGIVVEQDRPAGVNDKVRW
YCQKCQTVIHEVEFYLTDLGTQIKEAIVKFDADLDARTCKNCGTVNSSRRD
>O13287|Candida_albicans
MKNFNALSRLSILSKQLSFNNTNSSIARGDIGLIGLAVMGQNLILNMADHGYTVVAYNRT
TAKVDRFLENEAKGKSILGAHSIKELVDQLKRPRRIMLLVKAGAPVDEFINQLLPYLEEG
DIIIDGGNSHFPDSNRRYEELAKKGILFVGSGVSGGEEGARTGPSLMPGGNEKAWPHIKE
IFQDVAAKSDGEPCCDWVGDAGAGHYVKMVHNGIEYGDMQLICEAYDLMKRVGKFEDKEI
GDVFATWNKGVLDSFLIEITRDILYYNDPTDGKPLVEKILDTAGQKGTGKWTAVNALDLG
IPVTLIGEAVFSRCLSAMKAERVEASKALKGPQVTGESPITDKKQFIDDLEQALYASKII
SYTQGFMLMNQAAKDYGWKLNNAGIALMWRGGCIIRSVFLAEITAAYRKKPDLENLLLYP
FFNDAITKAQSGWRASVGKAIQYGIPTPAFSTALAFYDGLRSERLPANLLQAQRDYFGAH
TFKVLPGQENELLKKDEWIHINWTGRGGDVSSTTYDA
...
The site scansite.mit.edu provides a service that searches for motifs within proteins that are likely to be phosphorylated by specific protein kinases or bind to domains such as SH2 domains, 14-3-3 domains or PDZ domains.
The extract_scansite_motifs_fasta.pl script searches a given FASTA file (comprising protein sequences) against the scansite server and outputs a tab-delimited file containing all the significant motifs.
For example, you can run the script on Acetyl_CoA_hydrolase.fasta generated previously as:
[uzi@quince-srv2 ~/annotation]$ perl extract_scansite_motifs_fasta.pl -f Acetyl_coa_hydrolase.fasta
ID Kinase_name Percentile Position_in_protein Score Sequence Phosphorylated_residue Zscore
Q754Q2|Ashbya_gossypii Amphi_SH3 0.102 195 0.4737 GLHDIDMPVLPPHRV P195 -4.262
Q754Q2|Ashbya_gossypii CapC_SH3 0.076 222 0.5588 LDAVPVDPARVVALV P222 -4.497
P83773:Q59YK9|Candida_albicans PKC_epsilon 0.054 492 0.3526 FYATKKKTLHEPHIL T492 -3.338
P83773:Q59YK9|Candida_albicans ErkDD 0.110 216 0.5485 FKIGKTAIPVDPEKV I216 -3.583
Q6FPF3|Candida_glabrata Amphi_SH3 0.102 195 0.4737 GVHDIDMPVNPPHRV P195 -4.262
Q6FPF3|Candida_glabrata PDK1_Bind 0.057 340 0.3495 ERVFKNWDFYKSRLC D340 -5.194
Q6BKW1|Debaryomyces_hansenii Grb2_SH2 0.121 333 0.4046 EAGFDKFYENWDTYS Y333 -4.345
Q6BKW1|Debaryomyces_hansenii ErkDD 0.068 215 0.5206 YRTGLTAIPVDPSKV I215 -3.777
Q54K91|Dictyostelium_discoideum PKA_Kin 0.055 21 0.2758 ESRIKRKSLLSKVVT S21 -4.455
P15937:Q7RV87|Neurospora_crassa PKC_epsilon 0.139 410 0.3816 FLRNSKYSIMHTPST S410 -3.066
P15937:Q7RV87|Neurospora_crassa PKC_zeta 0.036 18 0.4109 RARVKRPSMLKKLCN S18 -3.778
P15937:Q7RV87|Neurospora_crassa PKC_mu 0.165 519 0.4187 KALVEEGSMAKVKF* S519 -3.263
P15937:Q7RV87|Neurospora_crassa ErkDD 0.086 221 0.5357 DRIGTTSVPVDPEKV V221 -3.672
Q9UUJ9:P78872|Schizosaccharomyces_pombe PDK1_Bind 0.040 339 0.2255 EKFYSNYDYYKERIL D339 -5.843
Q6C3Z9|Yarrowia_lipolytica ErkDD 0.117 144 0.5519 KKENSLDIAVIEATA I144 -3.559
PROKKA is a software tool for the rapid annotation of prokaryotic genomes. You can use it to generate GFF3, GBK and SQN files for metagenomic whole-genome shotgun contigs assembled using any metagenomic assembly software.
After identifying CDS regions using PROKKA, you can search the protein sequences against NCBI's Conserved Domain Databases (CDD).
CDD is a protein annotation resource that consists of a collection of well-annotated multiple sequence alignment models for ancient domains and full-length proteins. These are available as position-specific score matrices (PSSMs) for fast identification of conserved domains in protein sequences via RPS-BLAST.
While searching against these databases, refer to standard operating procedures for parameter selection.
You can use the PROKKA_RPSBLAST.sh script that has the following syntax (-d switch to specify the database):
[uzi@quince-srv2 ~/annotation]$ ./PROKKA_RPSBLAST.sh
Usage:
bash PROKKA_RPSBLAST.sh -f <fasta_file.faa> [options]
Options:
-p Parallelize using GNU Parallel flag
-c Number of cores to use (Default: 10)
-d rpsblast db [Cdd,Cog,Kog,Pfam,Prk,Smart,Tigr] to use (Default: Cog)
-t Number of threads to use (Default: 1)
-r Number of reference matches (Default: 500)
-e Expectation value (E) threshold for saving hits (Default: 0.00001)
For different databases, you can run the script as
./PROKKA_RPSBLAST.sh -p -c 20 -f PROKKA_XXXXXXXX/PROKKA_XXXXXXXX.faa -d Cdd
./PROKKA_RPSBLAST.sh -p -c 20 -f PROKKA_XXXXXXXX/PROKKA_XXXXXXXX.faa -d Cog
./PROKKA_RPSBLAST.sh -p -c 20 -f PROKKA_XXXXXXXX/PROKKA_XXXXXXXX.faa -d Kog
./PROKKA_RPSBLAST.sh -p -c 20 -f PROKKA_XXXXXXXX/PROKKA_XXXXXXXX.faa -d Pfam
./PROKKA_RPSBLAST.sh -p -c 20 -f PROKKA_XXXXXXXX/PROKKA_XXXXXXXX.faa -d Prk
./PROKKA_RPSBLAST.sh -p -c 20 -f PROKKA_XXXXXXXX/PROKKA_XXXXXXXX.faa -d Smart
./PROKKA_RPSBLAST.sh -p -c 20 -f PROKKA_XXXXXXXX/PROKKA_XXXXXXXX.faa -d Tigr
Cdd has all databases combined Cog/Kog/Pfam/Prk/Smart/Tigr i.e. everything. The PROKKA_CDD.py script can then be used with the GFF file generated from PROKKA:
[uzi@quince-srv2 ~/annotation]$ ./PROKKA_CDD.py
Usage:
./PROKKA_CDD.py -g <gfffile> -b <blastoutfile>
Optional parameters:
-s (--scovs-threshold) subject coverage threshold (Default:60)
-p (--pident-threshold) pident threshold (Default:0)
Example usage:
Step 1: Run PROKKA_XXXXXXXX.faa with rpsblast against the [Cdd/Cog/Kog/Pfam/Prk/Smart/Tigr] database
with following format:
rpsblast -query PROKKA_XXXXXXXX.faa -db [Cdd/Cog/Kog/Pfam/Prk/Smart/Tigr] -evalue 0.00001
-outfmt "6 qseqid sseqid evalue pident score qstart qend
sstart send length slen" -out blast_output.out
Step 2: Run this script to generate CDD anotations:
./PROKKA_CDD.py -g PROKKA_XXXXXXXX.gff -b blast_output.out
> annotations.tsv
The output is a tab-delimited file with following fields:
#Query with concatenated PROKKA CDS Ids using _ as delimeter
Hit from CDD database using NCBI's eutils
E-value from rpsblast
Identity from rpsblast
Score from rpsblast
Query-start from rpsblast
Query-end from rpsblast
Hit-start from rpsblast
Hit-end from rpsblast
Hit-length from rpsblast
Abstract from CDD database using NCBI's eutils
Title from CDD database using NCBI's eutils
Comments CDS to contig mapping (from GFF file)
Refer to rpsblast tutorial: http://www2.warwick.ac.uk/fac/sci/moac/people/students/peter_cock/python/rpsblast/
Here is the output generated on an example dataset:
[uzi@quince-srv2 ~/annotation]$ ./PROKKA_CDD.py -g ../PROKKA_01092014/PROKKA_01092014.gff -b PROKKA_01092014.out
#Query Hit E-value Identity Score Query-start Query-end Hit-start Hit-end Hit-length Abstract TitleComments
contig-9000000_07199 cd01189 1e-40 30.85 366 177 354 3 185 188 phiLC3 phage and phage-related integrases, site-specific recombinases, DNA breaking-rejoining enzymes, C-terminal catalytic domain. This CD includes various bacterial (mainly gram positive) and phage integrases, including those similar to Lactococcus phage phiLC3, TPW22, Tuc2009, BK5-T, A2, bIL285, bIL286, bIL311, ul36 and phi g1e; Staphylococcus aureus phage phi13 and phi42; Oenococcus oeni phage fOg44; Streptococcus thermophilus phage O1205 and Sfi21; and Streptococcus pyogenes phage T12 and T270. INT_phiLC3_C [138203,139355](-)
contig-9000000_07199 pfam00589 7e-25 29.19 250 177 356 3 167 185 Members of this family cleave DNA substrates by a series of staggered cuts, during which the protein becomes covalently linked to the DNA through a catalytic tyrosine residue at the carboxy end of the alignment. The catalytic site residues in CRE recombinase are Arg-173, His-289, Arg-292 and Tyr-324. Phage_integrase [138203,139355](-)
contig-9000000_07199 cd00801 8e-25 23.42 259 41 364 57 351 333 Bacteriophage P4 integrase. P4-like integrases are found in temperate bacteriophages, integrative plasmids, pathogenicity and symbiosis islands, and other mobile genetic elements. They share the same fold in their catalytic domain and the overall reaction mechanism with the superfamily of DNA breaking-rejoining enzymes. The P4 integrase mediates integrative and excisive site-specific recombination between two sites, called attachment sites, located on the phage genome and the bacterial chromosome. The phage attachment site is often found adjacent to the integrase gene, while the host attachment sites are typically situated near tRNA genes. INT_P4 [138203,139355](-)
...
KNOWN BUG WITH PROKKA AND RESOLUTION: PROKKA (verson 1.7.2) doesn't seem to handle longer contig names (from assembly software) in the FASTA file and messes up the GBK file after annotation. Before using PROKKA, you can change the FASTA headers using the following one-liner:
[uzi@quince-srv2 ~/annotation]$ awk '/>/{gsub("^>","",$0);split($0,a,"_");$0=">C"a[2]}1' test.fasta > test_mname.fasta
[uzi@quince-srv2 ~/annotation]$ grep -i ">" test.fasta | head
>NODE_3_length_194_cov_3.484536
>NODE_4_length_167_cov_3.233533
>NODE_5_length_462_cov_13.017316
>NODE_6_length_181_cov_2.651934
>NODE_7_length_171_cov_3.040936
>NODE_8_length_395_cov_3.055696
>NODE_9_length_244_cov_2.434426
>NODE_10_length_186_cov_2.951613
>NODE_11_length_212_cov_3.089623
>NODE_12_length_182_cov_3.307692
[uzi@quince-srv2 ~/annotation]$ grep -i ">" test_mname.fasta | head
>C3
>C4
>C5
>C6
>C7
>C8
>C9
>C10
>C11
>C12
Even if you have errors in your GBK file, you can still fix them. First of all, dates should be in the LOCUS line (grep -i "^LOCUS" on GBK file). In the problematic GBK file, some dates go down to the next line.
for example,
LOCUS NODE_100_length_1535_cov_6.6912051605 bp DNA linear
06-FEB-2014
DEFINITION Genus species strain strain.
Therefore, first step is to move all those lines up to the LOCUS line. You can save the file as test.gbk, edit it in vim, and use the following command (use the date that appears in your GBK file):
:%s/\n06-FEB-2014/ 06-FEB-2014/g
Secondly, notice that the length field is missing i.e. it says bp but where is the length? To resolve this, shorten the names and extract the lengths from contig names themselves i.e. by using the following command, "NODE_100_length_1535_cov_6.6912051605 bp" will change to "NODE_100 1535 bp" using this command:
[uzi@quince-srv2 ~/annotation]$ cat test.gbk | sed -e 's/\(NODE_[0-9]*\)_length_\([0-9]*\)_.* bp/\1\t\2 bp/g' > test2.gbk
If you load the GBK file up in Biopython, it will still throw warning messages. Why? because lengths extracted from the names are different than the actual lengths of the sequences!
/usr/lib64/python2.6/site-packages/biopython-1.62-py2.6-linux-x86_64.egg/Bio/GenBank/__init__.py:1191: BiopythonParserWarning: Expected sequence length 16779, found 16849 (NODE_97).
BiopythonParserWarning)
/usr/lib64/python2.6/site-packages/biopython-1.62-py2.6-linux-x86_64.egg/Bio/GenBank/__init__.py:1191: BiopythonParserWarning: Expected sequence length 5819, found 5889 (NODE_98).
BiopythonParserWarning)
/usr/lib64/python2.6/site-packages/biopython-1.62-py2.6-linux-x86_64.egg/Bio/GenBank/__init__.py:1191: BiopythonParserWarning: Expected sequence length 197, found 267 (NODE_99).
BiopythonParserWarning)
/usr/lib64/python2.6/site-packages/biopython-1.62-py2.6-linux-x86_64.egg/Bio/GenBank/__init__.py:1191: BiopythonParserWarning: Expected sequence length 28909, found 28979 (NODE_9)
Don't worry, the file will still be loaded in Biopython with corrected lengths. After loading in Biopython, write the records to a different file and it will fix the lengths error:
from Bio import SeqIO
input_handle = open("test2.gbk", "rU")
output_handle = open("test3.gbk", "w")
sequences = SeqIO.parse(input_handle, "genbank")
count = SeqIO.write(sequences, output_handle, "genbank")
output_handle.close()
input_handle.close()
For example, the original test2.gbk contains:
LOCUS NODE_100 1535 bp DNA linear 06-FEB-2014
DEFINITION Genus species strain strain.
ACCESSION
VERSION
KEYWORDS .
SOURCE Genus species
ORGANISM Genus species
Unclassified.
COMMENT Annotated using prokka 1.7.2 from http://www.vicbioinformatics.com.
FEATURES Location/Qualifiers
source 1..1605
/organism="Genus species"
/mol_type="genomic DNA"
/strain="strain"
ORIGIN
1 aaagtaaatt agggatattc agtaaatagt ataaaattag ccaactaatt catacacaaa
61 atgttgcgaa ttagttggct tttttgtatc tttaggtatt cccccgcaaa taaggtttgg
121 aatccaaaac gggggaaatt ccccaacaaa gatatggcta agatacaaaa tatttcggaa
181 attcacccta ctttgggctt tacagaattt gatattatag aaaaatatcg caagagtttt
241 catgagagtg agcttggcag gcttcattcg gtgtttccgt ttgagcgtat ggcaaagacc
301 atgggcctgt cggaacagcg actgggacgc aggaacatct tcagtccttc ggcgaagatc
361 gcccttatgg tcctgaaggc gtacaccgga ttctctgaca ggcagctggt ggaacatctt
421 aacgggaaca tacactacca gatgttctgc gggattatga taaatccgtc ctttcccata
481 accaactaca agatagtcag tgccatccgc aatgagatag cgtcccgtct tgatgttgat
541 tccctccagg aagtgctggc ttcacactgg aaaccctatc ttgacagcct tcacgtctgt
601 atgaccgatg ccacatgcta tgagagccat atgcgttttc ctacggatat gaaactcctt
661 tgggaaagtc tggaatggct ctacaggcaa atctgccttc attgcagaga tttgggtata
721 aggcgtccgc gcaacaagta tgcggatgtg gcgaagtcct atctgtccta ctgcaagaag
781 agaaagagga aggcttcaag gacaaggatg ctcaagcgtc gtatgatcag actccttgaa
841 aagctcctca tacagaggga tgaaatccat agagaacatg gaacctcact ccgatatacc
901 caggattacc agaagcgtct ttccatcatc agaaaggttc ttgtacagga aaaggagttg
961 tttgaaggca ggaaggtcag ggaccgcatc gtcagcattg accgtcatta tgtacgtccc
1021 attgtaagag gcaaggaaac annnnnnnnn nnnnnnnnnn nnnnnnnnnn nnnnnnnnnn
1081 nnnnnnnnnn nnnnnnnnnn nnnnnnnnnn nnnnnnnnnn nnnnnnnnnn nnnnnnnnnn
1141 nnnnnnnnnn nnnnnnnnnn nnnnnnnnnn nnnnnnnnnn nnnnnnnnnn nnnnnnnnnn
1201 nnnnnnnnnn nnnnnnnnnn nnnnnnnnnn nnnncaaaag aaagggccac ccgacttgaa
1261 ggcagcttcg gcacacagaa gcagcattac tcactctcaa ggataaaggc acggaacagg
1321 aagacggaaa tcctatggat tttctttgga atacatacgg caaatgccat actgatgata
1381 gataaaatca ggaaccgggc ggataaagcg gcatgacata aatttaccga cagaaccaga
1441 aggggtcatc agacctcttc cgaagcgtcg tgtctaacaa gataggatta tgtgaaaaaa
1501 cacagaaaaa gggcaataat aatggcatat aaagtgatta tactctatca actttatatg
1561 ccatcttact ttttagggac atttactgaa tatccctatt taaat
//
where as test3.gbk now contains the correct lengths:
LOCUS NODE_100 1605 bp DNA UNK 06-FEB-2014
DEFINITION Genus species strain strain.
ACCESSION NODE_100
VERSION NODE_100
KEYWORDS .
SOURCE Genus species
ORGANISM Genus species Unclassified.
.
COMMENT Annotated using prokka 1.7.2 from http://www.vicbioinformatics.com.
FEATURES Location/Qualifiers
source 1..1605
/strain="strain"
/mol_type="genomic DNA"
/organism="Genus species"
ORIGIN
1 aaagtaaatt agggatattc agtaaatagt ataaaattag ccaactaatt catacacaaa
61 atgttgcgaa ttagttggct tttttgtatc tttaggtatt cccccgcaaa taaggtttgg
121 aatccaaaac gggggaaatt ccccaacaaa gatatggcta agatacaaaa tatttcggaa
181 attcacccta ctttgggctt tacagaattt gatattatag aaaaatatcg caagagtttt
241 catgagagtg agcttggcag gcttcattcg gtgtttccgt ttgagcgtat ggcaaagacc
301 atgggcctgt cggaacagcg actgggacgc aggaacatct tcagtccttc ggcgaagatc
361 gcccttatgg tcctgaaggc gtacaccgga ttctctgaca ggcagctggt ggaacatctt
421 aacgggaaca tacactacca gatgttctgc gggattatga taaatccgtc ctttcccata
481 accaactaca agatagtcag tgccatccgc aatgagatag cgtcccgtct tgatgttgat
541 tccctccagg aagtgctggc ttcacactgg aaaccctatc ttgacagcct tcacgtctgt
601 atgaccgatg ccacatgcta tgagagccat atgcgttttc ctacggatat gaaactcctt
661 tgggaaagtc tggaatggct ctacaggcaa atctgccttc attgcagaga tttgggtata
721 aggcgtccgc gcaacaagta tgcggatgtg gcgaagtcct atctgtccta ctgcaagaag
781 agaaagagga aggcttcaag gacaaggatg ctcaagcgtc gtatgatcag actccttgaa
841 aagctcctca tacagaggga tgaaatccat agagaacatg gaacctcact ccgatatacc
901 caggattacc agaagcgtct ttccatcatc agaaaggttc ttgtacagga aaaggagttg
961 tttgaaggca ggaaggtcag ggaccgcatc gtcagcattg accgtcatta tgtacgtccc
1021 attgtaagag gcaaggaaac annnnnnnnn nnnnnnnnnn nnnnnnnnnn nnnnnnnnnn
1081 nnnnnnnnnn nnnnnnnnnn nnnnnnnnnn nnnnnnnnnn nnnnnnnnnn nnnnnnnnnn
1141 nnnnnnnnnn nnnnnnnnnn nnnnnnnnnn nnnnnnnnnn nnnnnnnnnn nnnnnnnnnn
1201 nnnnnnnnnn nnnnnnnnnn nnnnnnnnnn nnnncaaaag aaagggccac ccgacttgaa
1261 ggcagcttcg gcacacagaa gcagcattac tcactctcaa ggataaaggc acggaacagg
1321 aagacggaaa tcctatggat tttctttgga atacatacgg caaatgccat actgatgata
1381 gataaaatca ggaaccgggc ggataaagcg gcatgacata aatttaccga cagaaccaga
1441 aggggtcatc agacctcttc cgaagcgtcg tgtctaacaa gataggatta tgtgaaaaaa
1501 cacagaaaaa gggcaataat aatggcatat aaagtgatta tactctatca actttatatg
1561 ccatcttact ttttagggac atttactgaa tatccctatt taaat
//
Last Updated by Dr Umer Zeeshan Ijaz on 24/03/2014.



